| Content Disclaimer Copyright @2020. All Rights Reserved. |
StatsToDo: Generalized Linear Models
Links : Home Index (Subjects) Contact StatsToDo
|
Introduction Gaussian Binomial Multinomial Ordinal Poisson H I J K Negative Binomial The Generalized Linear Models (GLM) calculate regression coefficients relating a dependent variable to multiple independent variables. The value of the dependent variable is a linear combination of products of coefficient and value of each independent variable, so the model has the same appearence as that of multiple regression or covariance analysis. A particular advantage of the models is that the algorithms can cope with a combination of factors (group names in text) and values (numerical data), as well as dependent variables from a variety of probability distributions. This allows the models to be highly complex, providing the probability distribution of the data is correctly assumed. It is beyond the capability of this page to provide full explanation or web paged based program for GLM, as it is a vast subject, and the calculations complex. R code examples are offered on this page, as these algorithms are already fully developed, tested, and accepted. Only brief explanations, assisting users to negotiate the R codes are offered For those new to using R the page R_Exp provides an introduction and help for installation of the R packages For full explation and exploration of this area of statistics, users are referred to the references as the starting point. Users are also reminded to seek advice and assistance from experienced statisticians if they are new to this area of statistics. ProgramsThe panels of this page provides R codes for regression with the following distributions for the dependent variable
FormatEach program is described in one of the subsequent panels. Each panel containing an introduction and a program template, each in a sub-panelEach program is provided with a set of example data, to demonstrate the procedures. Please note:
ReferencesPlease note that the R code template contins only the minimum amount of code, and produces only the basic results. Users may want to produce a more complete program, including the many options that are availble with each procedure.References are provided in the Explanation panel of each program for this purpose
GLM: Dependent Variable with Gaussian (Normal) Distribution (Analysis of Variance)
Explanation
R Code
C
D
E
This panel provides explanations and example R codes for one of the procedures in Generalized Linear Models (GLM), where the dependent variable is a measurement that is normally distributed (Gaussian). The Gaussian model is sometimes termed General Linear Model, and should not be confused with Generalized Linear Models which described a whole genre of regression methods in different panels of this page.
GLM for Gussian distributed values produce results that are nearly the same as that of the Least Square Methods (Regression ans Analysis of Variance). Instead of mathematically partitioning the variance into its components, GLM uses adaptive modelling, fitting the data using numerical approximations. In the GLM for Gaussian distribution model
Referenceshttps://rcompanion.org/rcompanion/e_04.html An R Companion for the Handbook of Biological Statistics by Salvatore S. Mangiafico. Chapter on analysis of covariance
The example data is computer genrated to demonstrate the computation. It represents a study of birthweight, where the independent variables are sex of the baby (Boy or Girl), ethnicity of the mother (French, German or Greek), and gestational age at birth (weeks), and the dependent variable the weight of the newborn in grams. There are 22 babies (rows) in this set of data. As the independent variables include both factors (sex and ethnicity) and values (gestation), the model is that of Analysis of Covariance
The first step is to place the data in a data frame
# Step 1: Data entry to dataframe
myDat = ("
Sex Ethn Gest BWt
Girl Greek 37 3048
Boy German 36 2813
Girl French 41 3622
Girl Greek 36 2706
Boy German 35 2581
Boy French 39 3442
Girl Greek 40 3453
Boy German 37 3172
Girl French 35 2386
Boy Greek 39 3555
Girl German 37 3029
Boy French 37 3185
Girl Greek 36 2670
Boy German 38 3314
Girl French 41 3596
Boy Greek 38 3312
Girl German 39 3200
Boy French 41 3667
Boy Greek 40 3643
Girl German 38 3212
Girl French 38 3135
Girl Greek 39 3366
")
myDataFrame <- read.table(textConnection(myDat),header=TRUE)
myDataFrame$Sex <- factor(myDataFrame$Sex) # enforce variable as factor
myDataFrame$Ethn <- factor(myDataFrame$Ethn) # enforce variable as factor
summary(myDataFrame)
The summary of the data is as follows
> summary(myDataFrame)
Sex Ethn Gest BWt
Boy :10 French:7 Min. :35.00 Min. :2386
Girl:12 German:7 1st Qu.:37.00 1st Qu.:3034
Greek :8 Median :38.00 Median :3206
Mean :38.05 Mean :3187
3rd Qu.:39.00 3rd Qu.:3450
Max. :41.00 Max. :3667
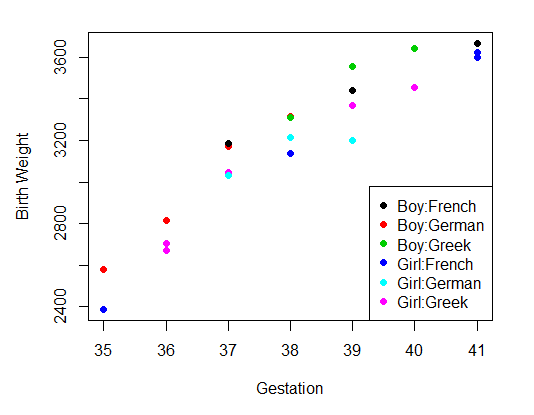
The second step can be plotting the data
# Step 2: Plot Data
sx <- factor(myDataFrame$Sex) #abbreviation
se <- factor(myDataFrame$Ethn) #abbreviation
par(pin=c(4.2, 3)) # set plotting window to 4.2x3 inches
plot(x = myDataFrame$Gest, # Gest on the x axis
y = myDataFrame$BWt, # BWt on the y axis
#col = factor(myDataFrame$Sex):factor(myDataFrame$Ethn), # in 6 colors (sex=2,Ethn=3)
col = sx:se, # in 6 colors (sex=2,Ethn=3)
pch = 16, # size of dot
xlab = "Gestation", # x label
ylab = "Birth Weight") # y lable
legend('bottomright', # placelegendon bottomright
legend = levels(sx:se), #all combinations
col = 1:6, # 6 colors for 6groups
cex = 1,
pch = 16)
Step 3a is optional, and performs analysis of covariance, including a test for interactions # Step 3a: Analysis of variance testing interactions testResults<-aov(BWt~Sex+Ethn+Gest+Sex*Ethn*Gest,data=myDataFrame) #Test for interactions summary(testResults) #show results Analysis of Variance TableThe results are in a table of analysis of variance from all sources, shown as follows
Df Sum Sq Mean Sq F value Pr(>F)
Sex 1 122427 122427 21.300 0.000958 ***
Ethn 2 278266 139133 24.206 0.000147 ***
Gest 1 2272527 2272527 395.376 2.27e-09 ***
Sex:Ethn 2 9208 4604 0.801 0.475688
Sex:Gest 1 4668 4668 0.812 0.388680
Ethn:Gest 2 8719 4360 0.759 0.493517
Sex:Ethn:Gest 2 71921 35960 6.256 0.017292 *
Residuals 10 57478 5748
The results of step 3a show significant interaction between Sex, Ethn and Gest. The interpretation is that different combinations of baby's sex and mother's ethnicity have different growth rates. If the data is real, and the sample size sufficiently large, then a more detailed analysis of this interaction is required before proceeding further. As this example data is computer generated, the sample size small, and the intention is to demonstrate the procedures, this significant interaction is ignored, and analysis will proceed to step 3b
Step 3b is a repeat of the analysis, but without the interaction terms # Step 3b: Final analysis of variance without interactions aovResults<-aov(BWt~Sex+Ethn+Gest,data=myDataFrame) #analysis of covariance summary(aovResults) #show results Analysis of Variance TableThe results are as follows
Df Sum Sq Mean Sq F value Pr(>F)
Sex 1 122427 122427 13.69 0.001776 **
Ethn 2 278266 139133 15.56 0.000144 ***
Gest 1 2272527 2272527 254.18 1.17e-11 ***
Residuals 17 151994 8941
This is a standard table of Analysis of variance. It shows that all 3 independent variables contributed significantly to values of birth weight.
If these results are accepted, then the next step, step 4, is to establich the formula where birthweight can be predicted from sex, maternal ethnicity and gestation age # Step 4: Generalized Linear Model formula glmResults<-lm(BWt~Sex+Ethn+Gest,data=myDataFrame) # Generalized Linear Model summary(glmResults) #show results Analysis of Variance TableThe results are
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -4021.50 467.16 -8.608 1.33e-07 ***
SexGirl -165.93 41.07 -4.040 0.000851 ***
EthnGerman 58.49 54.91 1.065 0.301682
EthnGreek 77.14 49.75 1.551 0.139430
Gest 190.61 11.96 15.943 1.17e-11 ***
The results to focus on is the column under the label Estimates, as this is the formula that models birth weight (BWt) according to Sex, Ethn, and Gestation
# Step 5: Check that formula works by calculating values and add to dataframe myDataFrame$Fitted<-fitted.values(glmResults) myDataFrameThe results are
Sex Ethn Gest BWt Fitted
1 Girl Greek 37 3048 2942.461
2 Boy German 36 2813 2899.123
3 Girl French 41 3622 3627.777
4 Girl Greek 36 2706 2751.846
5 Boy German 35 2581 2708.508
....
Step 6 is also optional. It pots the estimated birthweight against the actual bithweight, to show how closely the formula predicts
# Step 6: Plot original against calculated
par(pin=c(4.2, 3)) # set plotting window to 4.2x3 inches
plot(x = myDataFrame$BWt, # Bwt on the x axis
y = myDataFrame$Fitted, # Fit on the y axis
pch = 16, # size of dot
xlab = "Actual Birth Weight", # x label
ylab = "Calculated Birth Weight") # y lable
x_eq <- function(x){x} #curve function y=x
plot(x_eq, 2450, 3700, add=TRUE) #add curve to existing plot
The results are shown as follows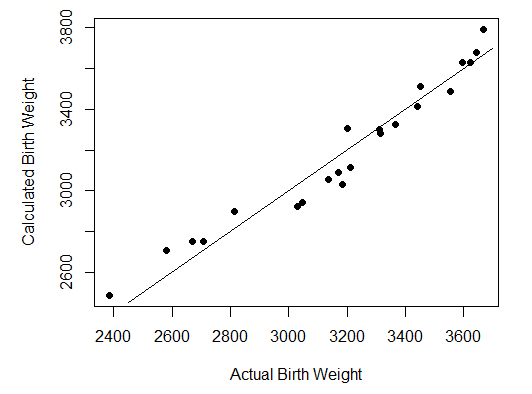
Step 7 demonstrate testing the formula on a new set of data. In 7a, a new data frame is created, consisting of the first 4 rows of the original data. In 7b, the formula produced in step 4 is then used to predict the birth weight
# Step 7a: create a new data frame
newDat = ("
Sex Ethn Gest
Girl Greek 37
Boy German 36
Girl French 41
Girl Greek 36
")
# create new dataframe from new input data
newDataFrame <- read.table(textConnection(newDat),header=TRUE)
summary(newDataFrame)
#step 7b: Estimate birthweight from established formula
newDataFrame$Model <- predict(glmResults, newdata=newDataFrame, type='response')
newDataFrame
The results are shown as follows
> summary(newDataFrame)
Sex Ethn Gest
Boy :1 French:1 Min. :36.0
Girl:3 German:1 1st Qu.:36.0
Greek :2 Median :36.5
Mean :37.5
3rd Qu.:38.0
Max. :41.0
>
> #step 7b:
> newDataFrame$Model <- predict(glmResults, newdata=newDataFrame, type='response')
> newDataFrame
Sex Ethn Gest Model
1 Girl Greek 37 2942.461
2 Boy German 36 2899.123
3 Girl French 41 3627.777
4 Girl Greek 36 2751.846
Step 7 demonstrate how the results of analysis (glmResults) can be used to estimate birth weight from a different data set. Please note the following
#Step 8: Optional saving and loading of coefficients
#save(glmResults, file = "glmResults.rda") #save results to rda file
#load("glmResults.rda") #load the rda file
The formula is saves as a .rda file, that can be reloaded on a different occasion and used on a different dataset. Please note the following
Contents of C:22
Contents of D:23
Contents of E:24
GLM: Dependent Variable with Binomial Distribution (Probability of a case belonging to one of two Groups)
Explanation
R Code
C
D
E
This page provides explanations and example R codes for Binomial Logistic Regression, where the dependent variable is the probabilities of belonging to one of the two groups.
GLM for binomial distribution is also known as Binomial Logistic Regression, or Logistic Regression for short. In this model, the dependent variable y is binomial, one of two outcomes, no/yes, True/false, 0/1. In R, the independent variables can be measurements or factors
The probability of outcome is then calculated by logistic transform p = 1 / (1 + exp(-y)) Referenceshttps://en.wikipedia.org/wiki/Logistic_regression Logistic Regression by Wikipedia Cox, DR (1958). "The regression analysis of binary sequences (with discussion)". J Roy Stat Soc B. 20 (2): 215 - 242. JSTOR 2983890. Portney LR, Watkins MP (2000) Foundations of Clinical Research Applications to Practice Second Edition.ISBN 0-8385-2695 0 p. 597 - 603
The example data is computer generated to demonstrate the computation. It perports to be from a study to predict the probability of Caesarean Section (factor of no/yes), based on the height of the mother (value), high blood pressure (factor no/yes), and presence of diabetes (factor 0_No, 1_Mild, 2_Severe)
Step 1 consists of creating the data frame from a data set or 5 columns and 22 births (rows)
# Step 1: Data entry
myDat = ("
Parity Diabetes HighBP Height CS
Multipara 1_Mild No 157 No
Nullipara 0_No Yes 157 No
Multipara 0_No No 153 Yes
Multipara 2_Severe No 165 No
Nullipara 0_No Yes 158 Yes
Nullipara 0_No No 158 Yes
Multipara 1_Mild No 151 Yes
Nullipara 0_No Yes 157 No
Multipara 0_No No 163 Yes
Nullipara 2_Severe No 163 Yes
Multipara 0_No Yes 162 No
Nullipara 0_No No 165 No
Multipara 1_Mild No 159 No
Nullipara 1_Mild Yes 159 Yes
Multipara 0_No No 157 Yes
Nullipara 2_Severe No 161 Yes
Multipara 0_No Yes 156 No
Nullipara 0_No No 159 Yes
Nullipara 1_Mild No 160 No
Multipara 0_No Yes 160 No
Multipara 0_No No 158 No
Multipara 2_Severe No 160 Yes
")
myDataFrame <- read.table(textConnection(myDat),header=TRUE)
summary(myDataFrame)
The summary of the data is as follows
> summary(myDataFrame)
Parity Diabetes HighBP Height CS
Multipara:12 0_No :13 No :15 Min. :151.0 No :11
Nullipara:10 1_Mild : 5 Yes: 7 1st Qu.:157.0 Yes:11
2_Severe: 4 Median :159.0
Mean :159.0
3rd Qu.:160.8
Max. :165.0
Step 2 is calculating the Binomial Logistic Regression
#Step 2: Binomial Logistic Regression binLogRegResults <- glm(CS~Parity+Diabetes+HighBP+Height,data=myDataFrame,family=binomial()) summary(binLogRegResults)The results are as follows
> summary(binLogRegResults)
Call:
glm(formula = CS ~ Parity + Diabetes + HighBP + Height, family = binomial(),
data = myDataFrame)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.25281 -0.89430 0.05138 0.62860 1.98546
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 60.2918 32.4084 1.860 0.0628 .
ParityNullipara 1.7912 1.2301 1.456 0.1454
Diabetes1_Mild -1.1484 1.3376 -0.859 0.3906
Diabetes2_Severe 2.0643 1.8111 1.140 0.2544
HighBPYes -2.0810 1.3889 -1.498 0.1340
Height -0.3811 0.2049 -1.860 0.0629 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 30.498 on 21 degrees of freedom
Residual deviance: 22.015 on 16 degrees of freedom
AIC: 34.015
Number of Fisher Scoring iterations: 5
The results show that none of the independent variables in this analysis has a statistically significant (p<0.05) influence on the outcome. This is not surprising as the data is computer generated and of a very small sample size.
The results to focus on are in the column under Estimates which are the coefficients. As an example we will use the fifth row of the data to demonstrate the math
#Step 3: Calculate probability of outcome using data myDataFrame$Fit<-fitted.values(binLogRegResults) #add predicted probability to dataframe myDataFrame #optional display of dataframeThe results are as follows, where the added column fit is the probability of yes for CS
> myDataFrame #optional display of dataframe
Parity Diabetes HighBP Height CS Fit
1 Multipara 1_Mild No 157 No 0.33558017
2 Nullipara 0_No Yes 157 No 0.54377606
3 Multipara 0_No No 153 Yes 0.87970051
4 Multipara 2_Severe No 165 No 0.37311861
5 Nullipara 0_No Yes 158 Yes 0.44880349
6 Nullipara 0_No No 158 Yes 0.86709292
7 Multipara 1_Mild No 151 Yes 0.83248000
8 Nullipara 0_No Yes 157 No 0.54377606
9 Multipara 0_No No 163 Yes 0.13931363
10 Nullipara 2_Severe No 163 Yes 0.88436769
11 Multipara 0_No Yes 162 No 0.02872213
12 Nullipara 0_No No 165 No 0.31175573
13 Multipara 1_Mild No 159 No 0.19074552
14 Nullipara 1_Mild Yes 159 Yes 0.14995209
15 Multipara 0_No No 157 Yes 0.61428430
16 Nullipara 2_Severe No 161 Yes 0.94249059
17 Multipara 0_No Yes 156 No 0.22537997
18 Nullipara 0_No No 159 Yes 0.81674315
19 Nullipara 1_Mild No 160 No 0.49124223
20 Multipara 0_No Yes 160 No 0.05959019
21 Multipara 0_No No 158 No 0.52106186
22 Multipara 2_Severe No 160 Yes 0.80002311
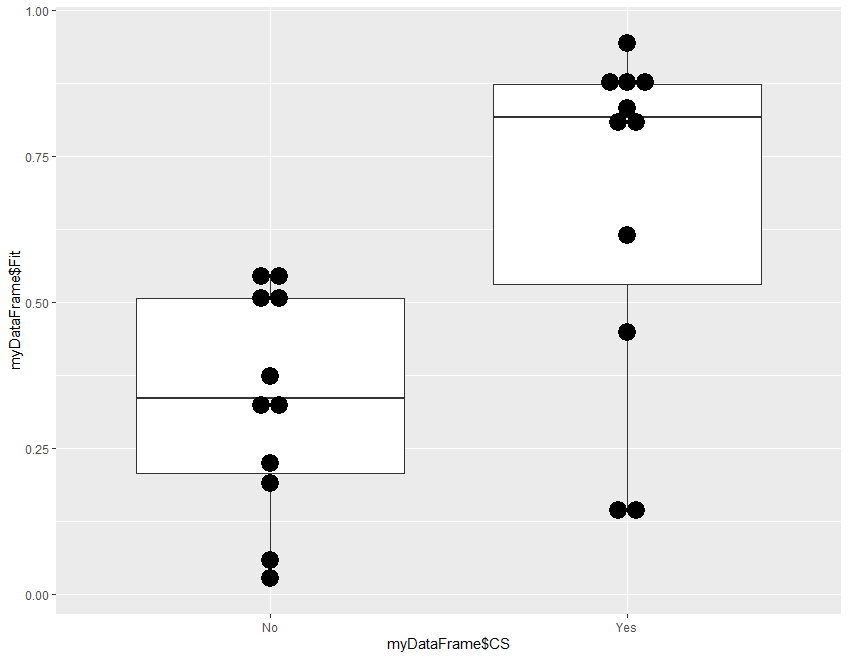
Step 4 is optional, a template for frequency plot of the probabilities in the two groups.
#step 4: Plot bar and dots of Fitted Probabilities
#install.packages("ggplot2") # if not already installed
library(ggplot2)
ggplot(myDataFrame, aes(x = myDataFrame$CS, y = myDataFrame$Fit)) +
geom_boxplot() +
geom_dotplot(binaxis="y",binwidth=.03,stackdir="center")
#Step 5a: Test results on a different set of data
newDat = ("
Parity Diabetes HighBP Height CS
Multipara 1_Mild No 157 No
Nullipara 0_No Yes 157 No
Multipara 0_No No 153 Yes
Multipara 2_Severe No 165 No
")
newDataFrame <- read.table(textConnection(newDat),header=TRUE)
summary(newDataFrame)
#Step 5b: test the model on new data frame
newDataFrame$Model <- predict(binLogRegResults, newdata=newDataFrame, type='response')
newDataFrame
The results are as follows
newDat = ("
+ Parity Diabetes HighBP Height CS
+ Multipara 1_Mild No 157 No
+ Nullipara 0_No Yes 157 No
+ Multipara 0_No No 153 Yes
+ Multipara 2_Severe No 165 No
+ ")
> newDataFrame <- read.table(textConnection(newDat),header=TRUE)
> summary(newDataFrame)
Parity Diabetes HighBP Height CS
Multipara:3 0_No :2 No :3 Min. :153 No :3
Nullipara:1 1_Mild :1 Yes:1 1st Qu.:156 Yes:1
2_Severe:1 Median :157
Mean :158
3rd Qu.:159
Max. :165
>
> newDataFrame$Model <- predict(binLogRegResults, newdata=newDataFrame, type='response')
> newDataFrame
Parity Diabetes HighBP Height CS Model
1 Multipara 1_Mild No 157 No 0.3355802
2 Nullipara 0_No Yes 157 No 0.5437761
3 Multipara 0_No No 153 Yes 0.8797005
4 Multipara 2_Severe No 165 No 0.3731186
Step 6 is to save the calculated model, and to load it when used subsequently
#Step 6: Optional saving and loading of coefficients
save(binLogRegResults, file = "binLogRegResults.rda") #save results to rda file
load("binLogRegResults.rda") #load the rda file
Step 6 is optional, and in fact commented out, and included as a template only. It allows the results of analysis to be stored as a .rda file, that can be reloaded on a different occasion and used on a different dataset. Please note the following
Contents of C:32
Contents of D:33
Contents of E:34
GLM: Dependent Variable with Multinomial Distribution (Probability of more than Two Groups)
Explanation
R Code
C
D
E
Multinomial Logistic Regression is an extension of the Binomial Logistic Regression, as explained in the previous panel, except that the dependent variable has more than two groups. The computation relies on the backpropagation neural network, one with no middle layer, and the transformation function is linear rather than logistic. The interpretation of results however are similar to other Generalized Linear Models In R, the independent variables can be measurements or factors
The algorithm produces m-1 formulae, m being the number of groups. The probabilities of belonging to the groups are estimated, and the estimated diagnosis assigned to that with the highest probability. Details on how this is carried out are described in the R code panel Referenceshttps://stats.idre.ucla.edu/r/dae/multinomial-logistic-regression/https://en.wikipedia.org/wiki/Multinomial_logistic_regression
The data is computer generated, and purports to be from a study of admissions from Emergency Department who felt unwell and has abdominal pain. The model is to make a diagnosis between appendicitis (Appen), Urinary Tract Infection (UTI), and the flu (Flu), based on the presence of vomiting (Vomit), the temperature (Temp) and pulse rate (Pulse).
Step 1 consists of creating the data frame from a data set or 4 columns and 15 cases (rows)
# Step 1: Data entry to dataframe
myDat = ("
Vomit Pulse Temp Diagnosis
No 74 102 Appen
Yes 68 97 UTI
No 78 99 Flu
Yes 72 98 UTI
No 72 101 Flu
Yes 92 96 Appen
No 76 100 UTI
No 86 100 Appen
No 74 98 Flu
No 72 99 UTI
Yes 84 100 Flu
Yes 85 98 Appen
No 64 103 Flu
No 78 97 UTI
Yes 72 99 Appen
")
myDataFrame <- read.table(textConnection(myDat),header=TRUE)
summary(myDataFrame)
The summary of the input data is shown as follows
> summary(myDataFrame)
Vomit Pulse Temp Diagnosis
No :9 Min. :64.00 Min. : 96.00 Appen:5
Yes:6 1st Qu.:72.00 1st Qu.: 98.00 Flu :5
Median :74.00 Median : 99.00 UTI :5
Mean :76.47 Mean : 99.13
3rd Qu.:81.00 3rd Qu.:100.00
Max. :92.00 Max. :103.00
Step 2:Performing Multinomial Regression
# Step 2: Multinomial Logistic Regression
#install.packages("nnet") # if not already installed
library(nnet)
mulLogRegResults <- multinom(formula = Diagnosis ~ Vomit + Pulse + Temp, data = myDataFrame)
summary(mulLogRegResults) #display results
The results are as follows
multinom(formula = Diagnosis ~ Vomit + Pulse + Temp, data = myDataFrame)
Coefficients:
(Intercept) VomitYes Pulse Temp
Flu 21.9547 -1.290127 -0.1272113 -0.1159828
UTI 143.5231 -2.036092 -0.3264960 -1.1910271
Std. Errors:
(Intercept) VomitYes Pulse Temp
Flu 0.02710540 1.641638 0.1015629 0.07724494
UTI 0.02030991 1.824531 0.1451480 0.10953534
Residual Deviance: 22.26156
AIC: 38.26156
The results to focus on are the rows under the heading Coefficients. As Appen is alphabetically the first group it is the reference group, and each row of the formulae produces the Y=log(Odds Ratio) for that diagnosis against that of appendicitis (Appen). The following shows how the calculations are done, using the first row of the data as an example (Vomit=No, Pulse=74, Temp=102)
In 3a, the procedure fitted produces the probabilities of the 3 diagnosis, and predict draws the conclusion to the most likely. User can then select the presentation preferred. In 3b, the computed diagnosis is compared with the diagnosis in the input data, to check the accuracies of the model. # Step 3a: Place fitted and predict values into data frame myFitted<-fitted.values(mulLogRegResults) myPredict <- predict(mulLogRegResults, myDataFrame) myDataFrame <- cbind(myDataFrame, myFitted, myPredict) myDataFrame # show resultsThe results are > myDataFrame # show results Vomit Pulse Temp Diagnosis Appen Flu UTI myPredict 1 No 74 102 Appen 0.31691544 0.64513376 0.037950792 Flu 2 Yes 68 97 UTI 0.02178191 0.04675669 0.931461399 UTI 3 No 78 99 Flu 0.25714354 0.44566188 0.297194581 Flu 4 Yes 72 98 UTI 0.17637011 0.20268118 0.620948707 UTI 5 No 72 101 Flu 0.21252412 0.62658442 0.160891453 Flu 6 Yes 92 96 Appen 0.85511578 0.09732217 0.047562044 Appen 7 No 76 100 UTI 0.27282910 0.54305543 0.184115473 Flu 8 No 86 100 Appen 0.63147967 0.35224218 0.016278145 Appen 9 No 74 98 Flu 0.05471871 0.17714340 0.768137895 UTI 10 No 72 99 UTI 0.07743112 0.28789115 0.634677730 UTI 11 Yes 84 100 Flu 0.83023903 0.16439342 0.005367549 Appen 12 Yes 85 98 Appen 0.78716931 0.17307872 0.039751967 Appen 13 No 64 103 Flu 0.11874659 0.76811862 0.113134788 Flu 14 No 78 97 UTI 0.06369797 0.13921848 0.797083550 UTI 15 Yes 72 99 Appen 0.32327816 0.33082179 0.345900049 UTIThe dataframe now contains the original columns, plus the 3 columns of probability for each diagnosis, as well as the conclusion which diagnosis is most likely. Now to check for accuracy # Step 3b. Check for accuracy with(myDataFrame, table(Diagnosis, myPredict)) #check table of prediction chisq.test(myDataFrame$Diagnosis, myDataFrame$myPredict)and the results are
myPredict
Diagnosis Appen Flu UTI
Appen 3 1 1
Flu 1 3 1
UTI 0 1 4
> chisq.test(myDataFrame$Diagnosis, myDataFrame$myPredict)
X-squared = 8.1, df = 4, p-value = 0.08798
The table shows the correct prediction along the diagonal, 10 cases out of 15, and 5 cases wrongly assigned. Chi Sq p=0.09 which is statistically insignificantly different from random distribution. This result is not surprising given the small size of sample, and computer generated values.
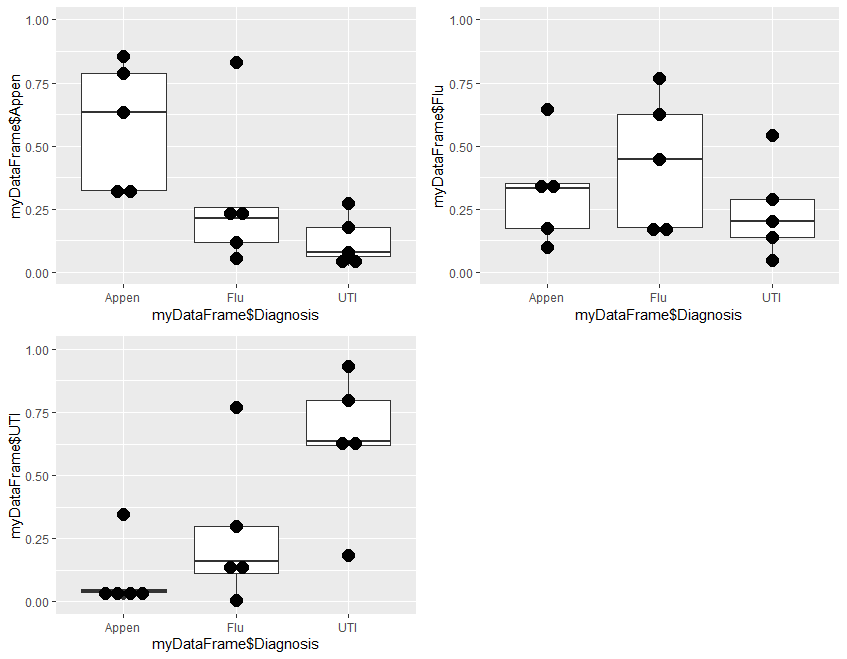
Step 4 is optional, and plots the results to visually demonstrate the results of analysis. It is divided into two parts. Step 4a produces 3 plots, and step 4b combines these 3 plots into a single one for presentation
# Step 4a: Plot 3 charts
#install.packages("ggplot2") # if not already installed
library(ggplot2)
plot1<-ggplot(myDataFrame, aes(x = myDataFrame$Diagnosis, y = myDataFrame$Appen)) +
coord_cartesian(ylim= c(0, 1)) +
geom_boxplot() + geom_dotplot(binaxis="y",binwidth=.05,stackdir="center") #fitted Appen probability
plot2<-ggplot(myDataFrame, aes(x = myDataFrame$Diagnosis, y = myDataFrame$UTI)) +
coord_cartesian(ylim= c(0, 1)) +
geom_boxplot() + geom_dotplot(binaxis="y",binwidth=.05,stackdir="center") #fitted Flu probability
plot3<-ggplot(myDataFrame, aes(x = myDataFrame$Diagnosis, y = myDataFrame$Flu)) +
coord_cartesian(ylim= c(0, 1)) +
geom_boxplot() + geom_dotplot(binaxis="y",binwidth=.05,stackdir="center") #fitted UTI probability
#Plot1
#plot2
#plot3
Plot1 plots the probability of Appen in the 3 groups of Appen, Flu, and UTI. Plot2 the probability of Flu and plot3 the probability of UTI in the same 3 groups.
At this stage, the 3 plots can be separately displayed. The problem is that, in RStudio, each plot over-writes the previous one, so the 3 cannot be visualized at the same time. To do this, we need to do 4b.
# Step 4b: Combine 3 charts into one
#install.packages("Grid") # only if not already installed
library(grid)
# Make a list from the ... arguments and plotlist
myPlots <- c(list(plot1, plot2, plot3)) #change list to your list of ggplots
myCols = 2 #change the number of plot columns per row of plots
# The rest of the codes need no change
myNumPlots = length(myPlots)
layout <- matrix(seq(1, myCols * ceiling(myNumPlots/myCols)),
ncol = myCols, nrow = ceiling(myNumPlots/myCols))
grid.newpage()
pushViewport(viewport(layout = grid.layout(nrow(layout), ncol(layout))))
# Make each plot, in the correct location
for (i in 1:myNumPlots) {
# Get the i,j matrix positions of the regions that contain this subplot
matchidx <- as.data.frame(which(layout == i, arr.ind = TRUE))
print(myPlots[[i]], vp = viewport(layout.pos.row = matchidx$row,
layout.pos.col = matchidx$col))
}
# Step 5a Use the coefficients on a new set of data
newDat = ("
Vomit Pulse Temp
No 74 102
Yes 68 97
No 78 99
")
newDataFrame <- read.table(textConnection(newDat),header=TRUE)
summary(newDataFrame)
$ Step 5b:
newProbs <- predict(mulLogRegResults, newdata=newDataFrame, type='probs')
newProbs # probabilies
newDiag <- predict(mulLogRegResults, newdata=newDataFrame, type='class')
newDiag # diagnosis
newDataFrame <- cbind(newDataFrame, newProbs, newDiag) # add to data frame
newDataFrame
The results are as follows
> summary(newDataFrame)
Vomit Pulse Temp
No :2 Min. :68.00 Min. : 97.00
Yes:1 1st Qu.:71.00 1st Qu.: 98.00
Median :74.00 Median : 99.00
Mean :73.33 Mean : 99.33
3rd Qu.:76.00 3rd Qu.:100.50
Max. :78.00 Max. :102.00
> newProbs
Appen Flu UTI
1 0.31691544 0.64513376 0.03795079
2 0.02178191 0.04675669 0.93146140
3 0.25714354 0.44566188 0.29719458
> newDiag
[1] Flu UTI Flu
Levels: Appen Flu UTI
>
> newDataFrame
Vomit Pulse Temp Appen Flu UTI newDiag
1 No 74 102 0.31691544 0.64513376 0.03795079 Flu
2 Yes 68 97 0.02178191 0.04675669 0.93146140 UTI
3 No 78 99 0.25714354 0.44566188 0.29719458 Flu
Please note the following
#Step 6: Optional saving and loading of coefficients
#save(mulLogRegResults, file = "MulLogRegResults.rda") #save results to rda file
#load("MulLogRegResults.rda") #load the rda file
Step 6 is optional, and in fact commented out, and included as a template only. It allows the results of analysis to be stored as a .rda file, that can be reloaded on a different occasion and used on a different dataset. Please note the following
Contents of C:42
Contents of D:43
Contents of E:44
GLM: Dependent Variable with Negative Binomial Distribution
Explanation
R Code
C
D
E
This panel provides explanations and example R codes for Negative Binomial Regression, where the dependent variable is a discrete positive integer with a negative binomial distribution.
Negative Binomial distribution is the reverse of the binomial distribution. Instead of stating a proportion, the statement is the number of one group is counted before a stated number of the other group is found. In Negative Binomial Regression, the algorithm is to estimate the number of one group found (failure, negative, 0, false) before finding each case of the other group (success, positive, 1, true), mostly counts of events or number of units. An examples is, instead of stating that the caesarean section rate is 20%, the Negative Binomial statement is having 4 vaginal deliveries for each Caesarean Section seen. Negative Binomial distribution is less rigid in its distribution assumptions than Poisson distribution, it is often adapted to analyse any measurements of discrete positive value where the Poisson distribution cannot be assured. The model Negative Binomial regression in the example on this page uses the same data as that in the Poisson panel. The results, other than minor rounding differences, are almost identical to the analysis assuming Poisson distribution. Negative Binomial regression, similar to other Generalized Linear Models, is conceptually and procedurally simple and easy to understand. The contraint however is that the count in the modelling data must be a positive integer, a value >0. When the expected count is low however, the count in some of the reference data may be zero, and this distorts the model. The remedy for this problem is to include a Zero Inflated Model in the algorithm, and this is explained in the R code panel Referenceshttps://en.wikipedia.org/wiki/Negative_binomial_distributionhttps://stats.idre.ucla.edu/r/dae/negative-binomial-regression/
The data is computer generated to demonstrate the computation methods. It perports to be from a study of workload in the labour ward, collected from a number of hospitals. Time is the time of the 3 shifts, AM for morning, PM for afternoon, and Night. Day is for day of the week, being Weekday or Weekend, BLY is the total number of births last year in thousands. The model is to make a estimate the number of births per shift (Births) from these 3 parameters, assuming that Births is a count and conforms to the Negative Binomial distribution
Step 1 is to create the data frame
# Step 1: Data entry
myDat = ("
Time Day BLY Births
PM Wday 4.5 6
PM Wend 5.6 4
Night Wday 6.0 3
PM Wday 5.9 8
Night Wday 5.2 3
AM Wend 5.8 4
Night Wend 5.9 7
AM Wend 5.0 0
Night Wday 5.5 4
Night Wday 4.1 6
PM Wday 5.4 3
Night Wday 4.2 5
PM Wday 4.7 3
AM Wday 4.4 4
AM Wend 4.7 2
Night Wend 4.8 5
PM Wend 4.8 6
PM Wday 4.5 5
AM Wday 5.0 9
")
myDataFrame <- read.table(textConnection(myDat),header=TRUE)
summary(myDataFrame)
The summary of input data is as follows
> summary(myDataFrame)
Time Day BLY Births
AM :5 Wday:12 Min. :4.100 Min. :0.000
Night:7 Wend: 7 1st Qu.:4.600 1st Qu.:3.000
PM :7 Median :5.000 Median :4.000
Mean :5.053 Mean :4.579
3rd Qu.:5.550 3rd Qu.:6.000
Max. :6.000 Max. :9.000
Strategy for Analysis
Firstly step 2a, using the glm model. #Step 2: Negative Binomial Regression #Step 2a. using glm #install.packages(MASS) library(MASS) glmResults<-glm.nb(formula = Births ~ Time + Day + BLY, data = myDataFrame) glmCount<-fitted.values(glmResults) #count = exp(y) summary(glmResults) myDataFrame <- cbind(myDataFrame, glmCount) #append glmCount to myDataFrame myDataFrameThe results are as follows
glm.nb(formula = Births ~ Time + Day + BLY, data = myDataFrame,
init.theta = 54599.73884, link = log)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.66322 -0.91210 -0.05635 0.47380 2.03454
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.27840 0.92893 1.376 0.169
TimeNight 0.15967 0.30042 0.531 0.595
TimePM 0.21978 0.29647 0.741 0.458
DayWend -0.16692 0.24589 -0.679 0.497
BLY 0.03089 0.18594 0.166 0.868
(Dispersion parameter for Negative Binomial(54599.74) family taken to be 1)
Null deviance: 21.614 on 18 degrees of freedom
Residual deviance: 20.151 on 14 degrees of freedom
AIC: 93.01
Number of Fisher Scoring iterations: 1
The results from Step 2a to focus on are the rows under the heading Estimate, as this is the formula for calculating the log(count). We will use the first row of data (Time=PM, Day=Wday, BLY=4.5) to demonstrate how this is done.
Time Day BLY Births glmCount
1 PM Wday 4.5 6 5.140811
2 PM Wend 5.6 4 4.500898
3 Night Wday 6.0 3 5.070469
4 PM Wday 5.9 8 5.368042
5 Night Wday 5.2 3 4.946685
6 AM Wend 5.8 4 3.635224
7 Night Wend 5.9 7 4.277745
8 AM Wend 5.0 0 3.546479
9 Night Wday 5.5 4 4.992746
10 Night Wday 4.1 6 4.781402
11 PM Wday 5.4 3 5.285758
12 Night Wday 4.2 5 4.796196
13 PM Wday 4.7 3 5.172674
14 AM Wday 4.4 4 4.113756
15 AM Wend 4.7 2 3.513761
16 Night Wend 4.8 5 4.134812
17 PM Wend 4.8 6 4.391018
18 PM Wday 4.5 5 5.140811
19 AM Wday 5.0 9 4.190723
>
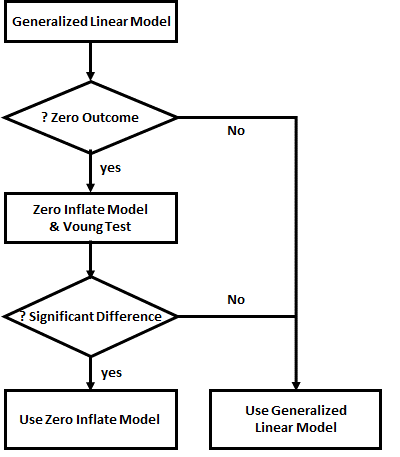
Next, in step 2b performs the zero inflated analysis, produces the output count accordingly, and append the results to the data frame.
The Vuong test compares the counts produced by the generalized linear model (glm) and zero inflated model (zi) and display whether the diffrence is significant.
#install.packages("pscl") # only if not previously installed
library(pscl)
ziResults<-zeroinfl(Births ~ Time + Day + BLY, dist="negbin", data = myDataFrame) #zero inlation model
summary(ziResults)
ziCount <- predict(ziResults, myDataFrame) #zero inflated count
myDataFrame <- cbind(myDataFrame,ziCount) #concatenate calculated count to myDataFrame
myDataFrame
The results are as follows
zeroinfl(formula = Births ~ Time + Day + BLY, data = myDataFrame, dist = "negbin")
Pearson residuals:
Min 1Q Median 3Q Max
-1.05117 -0.59996 -0.01863 0.49214 1.87321
Count model coefficients (negbin with log link):
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.51331 0.92356 1.639 0.101
TimeNight -0.01727 0.29613 -0.058 0.953
TimePM 0.04216 0.29228 0.144 0.885
DayWend -0.05730 0.24285 -0.236 0.813
BLY 0.01385 0.18458 0.075 0.940
Log(theta) 12.88701 167.64480 0.077 0.939
Zero-inflation model coefficients (binomial with logit link):
Estimate Std. Error z value Pr(>|z|)
(Intercept) -14.445 18942.295 -0.001 0.999
TimeNight -20.209 25664.897 -0.001 0.999
TimePM -20.310 25826.329 -0.001 0.999
DayWend 20.276 18942.290 0.001 0.999
BLY -1.281 3.020 -0.424 0.672
Theta = 395147.6678
Number of iterations in BFGS optimization: 24
Log-likelihood: -38.26 on 11 Df
> ziCount <- predict(ziResults, myDataFrame) #zero inflated count
> myDataFrame <- cbind(myDataFrame,ziCount) #concatenate calculated count to myDataFrame
> myDataFrame
Time Day BLY Births glmCount ziCount
1 PM Wday 4.5 6 5.140811 5.041842
2 PM Wend 5.6 4 4.500898 4.834124
3 Night Wday 6.0 3 5.070469 4.850651
4 PM Wday 5.9 8 5.368042 5.140529
5 Night Wday 5.2 3 4.946685 4.797217
6 AM Wend 5.8 4 3.635224 3.864226
7 Night Wend 5.9 7 4.277745 4.574173
8 AM Wend 5.0 0 3.546479 2.937645
9 Night Wday 5.5 4 4.992746 4.817185
10 Night Wday 4.1 6 4.781402 4.724706
11 PM Wday 5.4 3 5.285758 5.105064
12 Night Wday 4.2 5 4.796196 4.731252
13 PM Wday 4.7 3 5.172674 5.055823
14 AM Wday 4.4 4 4.113756 4.827030
15 AM Wend 4.7 2 3.513761 2.502474
16 Night Wend 4.8 5 4.134812 4.505033
17 PM Wend 4.8 6 4.391018 4.780872
18 PM Wday 4.5 5 5.140811 5.041842
19 AM Wday 5.0 9 4.190723 4.867299
Finally, in step 2c, the results from the Generalized Linear and Zero Inflated models are compared, using the Vuong Test. The Spearman's Correlation Coefficient is used for comparison
vuong(glmResults, ziResults)# comparing the two models cor.test( ~ as.numeric(Births) + as.numeric(glmCount), data=myDataFrame, method = "spearman",exact=FALSE) cor.test( ~ as.numeric(Births) + as.numeric(ziCount), data=myDataFrame, method = "spearman",exact=FALSE) cor.test( ~ as.numeric(glmCount) + as.numeric(ziCount), data=myDataFrame, method = "spearman",exact=FALSE)The results are as follows
>
> vuong(glmResults, ziResults)# comparing the two models
Vuong Non-Nested Hypothesis Test-Statistic:
(test-statistic is asymptotically distributed N(0,1) under the
null that the models are indistinguishible)
-------------------------------------------------------------
Vuong z-statistic H_A p-value
Raw -0.7669298 model2 > model1 0.22156
AIC-corrected 0.9437815 model1 > model2 0.17264
BIC-corrected 1.7516127 model1 > model2 0.03992
>
Spearman's rank correlation rho
> cor.test( ~ as.numeric(Births) + as.numeric(glmCount), data=myDataFrame, method = "spearman",exact=FALSE)
S = 976.91, p-value = 0.559, rho = 0.1430578
> cor.test( ~ as.numeric(Births) + as.numeric(ziCount), data=myDataFrame, method = "spearman",exact=FALSE)
S = 896.89, p-value = 0.3807 rho = 0.2132539
> cor.test( ~ as.numeric(glmCount) + as.numeric(ziCount), data=myDataFrame, method = "spearman",exact=FALSE)
data: as.numeric(glmCount) and as.numeric(ziCount)
S = 178.16, p-value = 5.647e-06, rho = 0.8437226
The results of the zero inflated analysis presents coefficients for combined Negative Binomial and Binmomial distributions to be used in estimating the counts. The method of calculation using these coefficients is by numerical approximation, and too complex to be explained here. Those interested in numerical methods should consult the references provided.
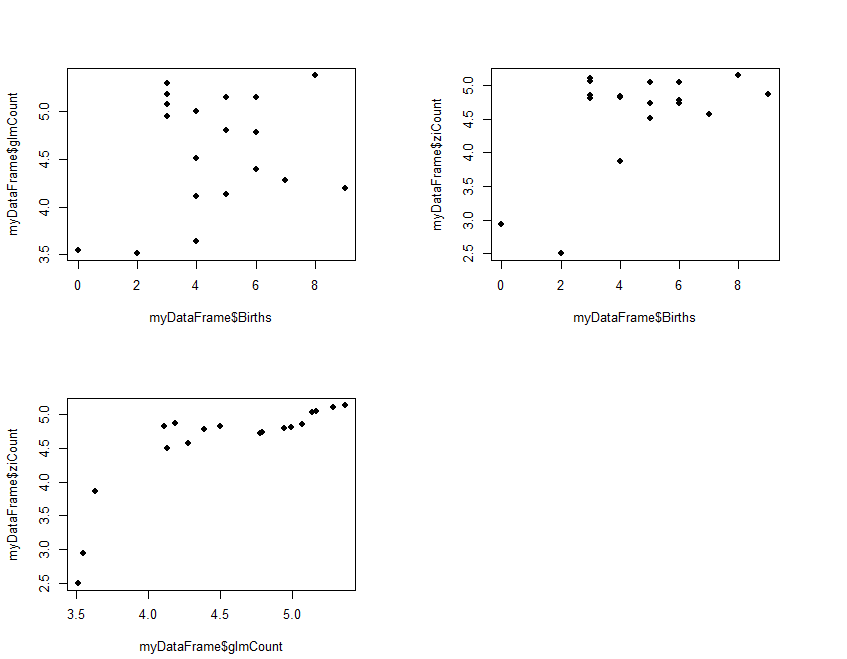
The results (ziCount) are then compared with those from the generalized linear model (glmCount) using the Voung test. The test produced by the basic calculation is labelled Raw, AIC has a correction by Akaike's information criterion, and BIC has a correction by the Bayesian information criterion. In practice they are different methods of calculating the same thing. One way of interpreting the 3 results is to assume that the two models are different if any of the 3 comparisons is statistically significant, as is the case with the current example data. The 3 Spearman's correlations test how closely the two estimates related to the original outcome, and to each other. The results shows that both performed poorly with the reference outcome (Births:glmCount, ρ=0.14; Births:ziCount, ρ=0.21; both not statistically significant). The two estimates however are closely but not perfectly correlated to each other (glmCount:ziCount, ρ=0.84, p<0.0001). Such poor results are not surprising, given that the data is randomly generated by the computer and the sample size very small. It merely serves to demonstrate the numerical methods. Step 3 is optional and plots the fitted values
#Step 3: Plot fitted values
old.par <- par(mfrow=c(2,2))
par(pin=c(3, 2)) # set plotting window to 4.2x3 inches
plot(x = myDataFrame$Births, # Births on the x axis
y = myDataFrame$glmCount, # glmCount on the y axis
xlim = c(min(myDataFrame$Births),max(myDataFrame$Births)),
ylim = c(min(myDataFrame$glmCount),max(myDataFrame$glmCount)),
pch = 16) # size of dot
plot(x = myDataFrame$Births, # Births on the x axis
y = myDataFrame$ziCount, # ziCount on the y axis
xlim = c(min(myDataFrame$Births),max(myDataFrame$Births)),
ylim = c(min(myDataFrame$ziCount),max(myDataFrame$ziCount)),
pch = 16) # size of dot
plot(x = myDataFrame$glmCount, # Births on the x axis
y = myDataFrame$ziCount, # ziCount on the y axis
xlim = c(min(myDataFrame$glmCount),max(myDataFrame$glmCount)),
ylim = c(min(myDataFrame$ziCount),max(myDataFrame$ziCount)),
pch = 16) # size of dot
par(old.par)
# Step 4a: Test coefficients on new data set
newDat = ("
Time Day BLY
PM Wday 4.5
Night Wday 6.0
AM Wend 5.8
")
newDataFrame <- read.table(textConnection(newDat),header=TRUE)
summary(newDataFrame)
# Step 4b: Calculate results using new data
newGlmCount <- predict(glmResults, newdata=newDataFrame, type='response')
newZiCount <- predict(ziResults, newdata=newDataFrame, type='response')
newDataFrame <- cbind(newDataFrame, newGlmCount, newZiCount)
newDataFrame
The results are as follows. Please note that, in practice, only one of the counts need to be estimated, as by this stage a decision whether glm or zi should be used has been made. The results are presented as follows
Time Day BLY newGlmCount newZiCount 1 PM Wday 4.5 5.140811 5.041842 2 Night Wday 6.0 5.070469 4.850651 3 AM Wend 5.8 3.635224 3.864226Please note the following
#Step 5: Optional saving and loading of coefficients
#save(glmResults, file = "NegBinGlmResults.rda") #save glm results to rda file
#load("NegBinGlmResults.rda") #load the glm rda file
#save(ziResults, file = "NegBinZiResults.rda") #save zi results to rda file
#load("NegBinZiResults.rda") #load the zi rda file
Please note that the codes are commented out as they are optional, and included as a template only. It allows the results of analysis to be stored as a .rda file, that can be reloaded on a different occasion and used on a different dataset. Please note the following
Contents of C:52
Contents of D:53
Contents of E:54
GLM: Dependent Variable with Ordinal Distribution
Explanation
R Code
C
D
E
This panel provides explanations and example R codes for Ordinal Logistic Regression, which is one of the algorithms based on the Generalized Linear Models. Ordinal Regression is a variant of the Multinomial Regression model, as explained in the previous panel, but the dependent variable is ordinal, in that they are ordered in scale alphabetically, so the grp1<Grp2<Grp3 ...
In R, the independent variables can be measurements or factors
Referenceshttps://en.wikipedia.org/wiki/Ordered_logithttps://stats.idre.ucla.edu/r/dae/ordinal-logistic-regression/
The data presented here are computer generated. It purports to be from a study on factors that predicts the success of breast feeding.
The predictors are whether they received antenatal breast feeding counselling (Counsel = No/Yes), level of education (Educ = Primary/Secondary/Tertiary), and age in completed years (Age). Outcomes are level success in breast feeding with the following levels (BFeed)
Step 1: Creates the data frame for analysis
# Step 1: Data entry to dataframe
myDat = ("
# Step 1: Data entry
myDat = ("
Counsel Educ Age BFeed
No Tertiary 27 S3
No Tertiary 29 S1
Yes Secondary 27 S1
No Primary 37 S2
No Tertiary 27 S0
No Primary 37 S3
Yes Secondary 25 S0
Yes Primary 33 S1
No Tertiary 35 S2
Yes Secondary 29 S1
No Primary 25 S0
Yes Tertiary 33 S2
Yes Secondary 27 S3
")
myDataFrame <- read.table(textConnection(myDat),header=TRUE)
summary(myDataFrame)
The summary of the data is as follows
> summary(myDataFrame)
Counsel Educ Age BFeed
No :7 Primary :4 Min. :25.00 S0:3
Yes:6 Secondary:4 1st Qu.:27.00 S1:4
Tertiary :5 Median :29.00 S2:3
Mean :30.08 S3:3
3rd Qu.:33.00
Max. :37.00
Step 2: Ordinal Regression
# Step 2: Ordinal Regression
#install.packages("MASS") # if not already installed
library(MASS)
ordRegResults <- polr(BFeed ~ Counsel + Educ + Age, data = myDataFrame, Hess=TRUE)
summary(ordRegResults)
The results are as follows
Call:
polr(formula = BFeed ~ Counsel + Educ + Age, data = myDataFrame,
Hess = TRUE)
Coefficients:
Value Std. Error t value
CounselYes -0.353 1.340 -0.263
EducSecondary 2.319 2.122 1.093
EducTertiary 1.229 1.371 0.896
Age 0.427 0.206 2.073
Intercepts:
Value Std. Error t value
S0|S1 12.36 6.75 1.83
S1|S2 14.52 7.19 2.02
S2|S3 15.87 7.35 2.16
Residual Deviance: 29.90
AIC: 43.90
The results from Step 2 to focus on are the rows under the heading Value in the two sets of Coefficients and Intercepts. We will use the first row of the data as an example of calculation (Counsel=No, Educ=Tertiary, Age=27)
The probabilities of each level are calculated as follows
In step 3a, the fitted.values function creates the 4 columns of probabilities for the levels of outcomes, and the predict function make the decision which outcome is the most likely. The cbind function concatenates these results to the dataframe, which is then displayed. Step 3b creates the 4x4 table of counts between the original Diagnosis and the estimated myPredict, to test the accuracy of the algorithm. The Spearman's Correlation Coefficient estimates the precision of the prediction (0=random, 1=perfect). # Step 3a: Place fitted values into data frame and display results myFitted<-round(fitted.values(ordRegResults),4) myPredict <- predict(ordRegResults, myDataFrame) myDataFrame <- cbind(myDataFrame, myFitted, myPredict) myDataFrame #optional display of data frameThe results are as follows Counsel Educ Age BFeed S0 S1 S2 S3 myPredict 1 No Tertiary 27 S3 0.4049 0.4502 0.1027 0.0423 S1 2 No Tertiary 29 S1 0.2247 0.4906 0.1908 0.0939 S1 3 Yes Secondary 27 S1 0.2455 0.4928 0.1772 0.0845 S1 4 No Primary 37 S2 0.0316 0.1889 0.3003 0.4792 S3 5 No Tertiary 27 S0 0.4049 0.4502 0.1027 0.0423 S1 6 No Primary 37 S3 0.0316 0.1889 0.3003 0.4792 S3 7 Yes Secondary 25 S0 0.4331 0.4357 0.0934 0.0378 S1 8 Yes Primary 33 S1 0.2037 0.4855 0.2058 0.1051 S1 9 No Tertiary 35 S2 0.0219 0.1408 0.2647 0.5726 S3 10 Yes Secondary 29 S1 0.1218 0.4241 0.2761 0.1780 S1 11 No Primary 25 S0 0.8451 0.1342 0.0152 0.0055 S0 12 Yes Tertiary 33 S2 0.0696 0.3239 0.3202 0.2863 S1 13 Yes Secondary 27 S3 0.2455 0.4928 0.1772 0.0845 S1 # Step 3b with(myDataFrame, table(BFeed,myPredict)) #check table of prediction cor.test( ~ as.numeric(BFeed) + as.numeric(myPredict), data=myDataFrame, method = "spearman",exact=FALSE)The results are
> with(myDataFrame, table(BFeed,myPredict)) #check table of prediction
myPredict
BFeed S0 S1 S2 S3
S0 1 2 0 0
S1 0 4 0 0
S2 0 1 0 2
S3 0 2 0 1
> cor.test( ~ as.numeric(BFeed) + as.numeric(myPredict), data=myDataFrame, method = "spearman",exact=FALSE)
Spearman's rank correlation rho
S = 200, p-value = 0.05, rho = 0.55
The dataframe now contains the original columns, plus the 4 columns of probability for each level of outcome, as well as the conclusion which level is most likely. The table shows the correct prediction along the diagonal, 6 cases out of 13. The Spearman's correlation coefficient is 0.55
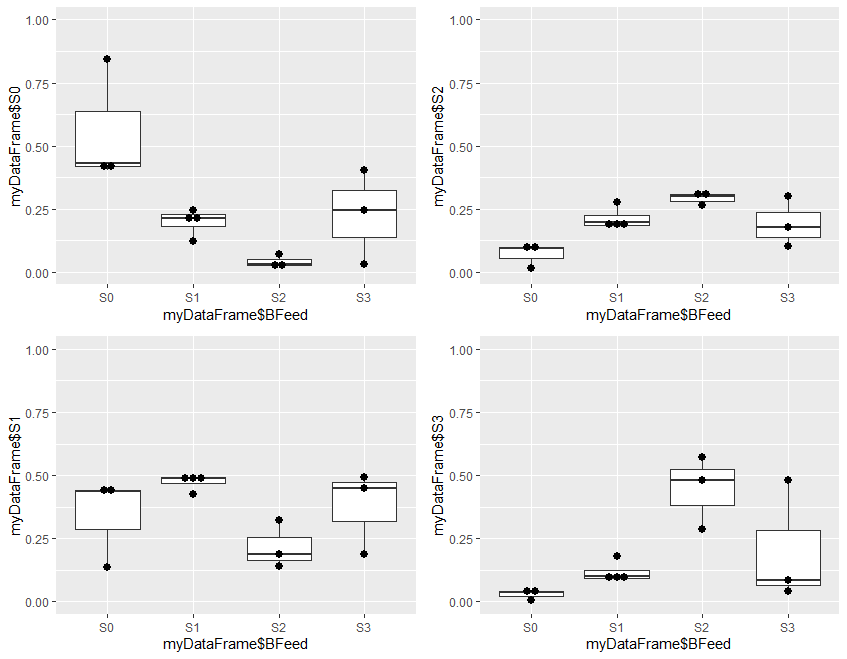
Step 4 is optional, and plots the results to visually demonstrate the results of analysis. It is divided into two parts. Step 4a produces 4 plots, and step 4b combines these 4 plots into a single one for presentation
#Step 4a: Make 4 different plots
#install.packages("ggplot2") # if not already installed
library(ggplot2)
plot1<-ggplot(myDataFrame, aes(x = myDataFrame$BFeed, y = myDataFrame$S0)) +
coord_cartesian(ylim= c(0, 1)) +
geom_boxplot() + geom_dotplot(binaxis="y",binwidth=.03, stackdir="center")
plot2<-ggplot(myDataFrame, aes(x = myDataFrame$BFeed, y = myDataFrame$S1, ymin=0.0, ymax=1.0)) +
coord_cartesian(ylim= c(0, 1)) +
geom_boxplot() + geom_dotplot(binaxis="y",binwidth=.03, stackdir="center")
plot3<-ggplot(myDataFrame, aes(x = myDataFrame$BFeed, y = myDataFrame$S2, ymin=0.0, ymax=1.0)) +
coord_cartesian(ylim= c(0, 1)) +
geom_boxplot() + geom_dotplot(binaxis="y",binwidth=.03, stackdir="center")
plot4<-ggplot(myDataFrame, aes(x = myDataFrame$BFeed, y = myDataFrame$S3, ymin=0.0, ymax=1.0)) +
coord_cartesian(ylim= c(0, 1)) +
geom_boxplot() + geom_dotplot(binaxis="y",binwidth=.03, stackdir="center")
#plot1
#plot2
#plot3
#plot4
Plot1 plots the estimated probability of S0 for the 4 levels of outcome in the original data, plot2 the probability of S1, plot3 the probability of S2, and plot4 the probability of S3.
At this stage, the 4 plots can be separately displayed. The problem is that, in RStudio, each plot over-writes the previous one, so the 4 cannot be visualized at the same time. To do this, we need to do step 4b.
# Step 4b: Combine 4 plots into 1
#install.packages("Grid") # if not already installed
library(grid)
# Make a list from the ... arguments and plotlist
myPlots <- c(list(plot1,plot2,plot3,plot4)) #change list to your list of ggplots
myCols = 2 #change the number of plot columns per row of plots
# The rest of the codes need no change
myNumPlots = length(myPlots)
layout <- matrix(seq(1, myCols * ceiling(myNumPlots/myCols)),
ncol = myCols, nrow = ceiling(myNumPlots/myCols))
grid.newpage()
pushViewport(viewport(layout = grid.layout(nrow(layout), ncol(layout))))
# Make each plot, in the correct location
for (i in 1:myNumPlots) {
# Get the i,j matrix positions of the regions that contain this subplot
matchidx <- as.data.frame(which(layout == i, arr.ind = TRUE))
print(myPlots[[i]], vp = viewport(layout.pos.row = matchidx$row,
layout.pos.col = matchidx$col))
}
# Step 5a: Creates a different set of data
newDat = ("
Counsel Educ Age
No Tertiary 27
No Tertiary 29
Yes Secondary 25
Yes Tertiary 33
")
newDataFrame <- read.table(textConnection(newDat),header=TRUE)
summary(newDataFrame)
#step 5b: Test coefficients on new set of data
newProbs <- predict(ordRegResults, newdata=newDataFrame, type='probs')
newProbs
newDiag <- predict(ordRegResults, newdata=newDataFrame, type='class')
newDiag
# Add results to new data frame
newDataFrame <- cbind(newDataFrame, newProbs, newDiag)
newDataFrame
The results are shown as follows
Counsel Educ Age S0 S1 S2 S3 newDiag 1 No Tertiary 27 0.40 0.45 0.103 0.042 S1 2 No Tertiary 29 0.22 0.49 0.191 0.094 S1 3 Yes Secondary 25 0.43 0.44 0.093 0.038 S1 4 Yes Tertiary 33 0.07 0.32 0.320 0.286 S1Please note the following
#Step 6: Optional saving and loading of coefficients
#save(ordRegResults, file = "OrdRegResults.rda") #save results to rda file
#load("OrdRegResults.rda") #load the rda file
Step 6 is optional, and in fact commented out, and included as a template only. It allows the results of analysis to be stored as a .rda file, that can be reloaded on a different occasion and used on a different dataset. Please note the following
Contents of C:62
Contents of D:63
Contents of E:64
GLM: Dependent Variable with Poisson Distribution (Counts)
Explanation
R Code
C
D
E
This page provides explanations and example R codes for Poisson Regression, where the dependent variable is a discrete positive integer with Poisson distribution, mostly counts of events or number of units in a defined environment. Examples are the number of cells seen in a field under the microscope, the number of asthma attacks per 100 child year, the number of pregnancies per 100 women year using a particular form of contraception.
The Poisson Regression is a powerful analytical tool, providing that the data conforms to the Poisson distribution. When there is doubt about the distribution, the less powerful methods with less rigorous assumptions should be used. These are the Ordinal Regression or the Negative Binomial Regression, as explained in the other panels of this page Poisson regression, similar to other Generalized Linear Models, is conceptually and procedurally simple and easy to understand. The contraint however is that the count in the modelling data must be a positive integer, a value >0. When the expected count is low however, the count in some of the reference data may be zero, and this distorts the model. The remedy for this problem is to include a Zero Inflated Model in the algorithm, and this is explained in the R code panel Referenceshttps://stats.idre.ucla.edu/r/dae/poisson-regression/ https://stats.idre.ucla.edu/r/dae/zip/ https://en.wikipedia.org/wiki/Zero-inflated_model https://en.wikipedia.org/wiki/Vuong%27s_closeness_test https://www.rdocumentation.org/packages/mpath/versions/0.1-20/topics/vuong.test
The data is computer generated to demonstrate the computation methods. It perports to be from a study of workload in the labour ward, collected from a number of hospitals. Time is the time of the 3 shifts, AM for morning, PM for afternoon, and Night. Day is for day of the week, being Weekday or Weekend, BLY is the total number of births last year in thousands. The model is to make a estimate the number of births per shift (Births) from these 3 parameters, assuming that Births is a count and conforms to the Poisson distribution
Step 1 creates the dataframe that contains the data as in myDat.
# Step 1: Data entry
myDat = ("
Time Day BLY Births
PM Wday 4.5 6
PM Wend 5.6 4
Night Wday 6.0 3
PM Wday 5.9 8
Night Wday 5.2 3
AM Wend 5.8 4
Night Wend 5.9 7
AM Wend 5.0 0
Night Wday 5.5 4
Night Wday 4.1 6
PM Wday 5.4 3
Night Wday 4.2 5
PM Wday 4.7 3
AM Wday 4.4 4
AM Wend 4.7 2
Night Wend 4.8 5
PM Wend 4.8 6
PM Wday 4.5 5
AM Wday 5.0 9
")
myDataFrame <- read.table(textConnection(myDat),header=TRUE)
summary(myDataFrame)
The summary of the input data is as follows
> summary(myDataFrame)
Time Day BLY Births
AM :5 Wday:12 Min. :4.100 Min. :0.000
Night:7 Wend: 7 1st Qu.:4.600 1st Qu.:3.000
PM :7 Median :5.000 Median :4.000
Mean :5.053 Mean :4.579
3rd Qu.:5.550 3rd Qu.:6.000
Max. :6.000 Max. :9.000
Strategy for Analysis
Step 2a performs the analysis using the glm model. The results are as follows #Step 2a. using glm glmResults<-glm(formula = Births ~ Time + Day + BLY, data = myDataFrame, family=poisson()) summary(glmResults) #display results glmCount<-fitted.values(glmResults) #count = exp(y) myDataFrame <- cbind(myDataFrame, glmCount) #append glmCount to myDataFrame myDataFrameThe results are as follows
Call:
glm(formula = Births ~ Time + Day + BLY, family = poisson(),
data = myDataFrame)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.66327 -0.91213 -0.05635 0.47382 2.03464
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.27841 0.92889 1.376 0.169
TimeNight 0.15966 0.30041 0.531 0.595
TimePM 0.21978 0.29646 0.741 0.458
DayWend -0.16691 0.24588 -0.679 0.497
BLY 0.03089 0.18593 0.166 0.868
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 21.615 on 18 degrees of freedom
Residual deviance: 20.153 on 14 degrees of freedom
AIC: 91.01
Number of Fisher Scoring iterations: 5
The results from Step 2a to focus on are the rows under the heading Estimate, as this is the formula for calculating the log(count). We will use the first row of data (Time=PM, Day=Wday, BLY=4.5) to demonstrate how this is done.
Time Day BLY Births glmCount
1 PM Wday 4.5 6 5.140809
2 PM Wend 5.6 4 4.500904
3 Night Wday 6.0 3 5.070451
4 PM Wday 5.9 8 5.368030
5 Night Wday 5.2 3 4.946672
6 AM Wend 5.8 4 3.635237
7 Night Wend 5.9 7 4.277743
8 AM Wend 5.0 0 3.546494
9 Night Wday 5.5 4 4.992731
10 Night Wday 4.1 6 4.781396
11 PM Wday 5.4 3 5.285749
12 Night Wday 4.2 5 4.796190
13 PM Wday 4.7 3 5.172670
14 AM Wday 4.4 4 4.113764
15 AM Wend 4.7 2 3.513777
16 Night Wend 4.8 5 4.134816
17 PM Wend 4.8 6 4.391029
18 PM Wday 4.5 5 5.140809
19 AM Wday 5.0 9 4.190728
Next, step 2b calculates the Zero-inflation, and adds the results to the data frame
#Step 2b. Zero-inflation
#install.packages("pscl") # only if not previously installed
library(pscl)
ziResults<-zeroinfl(Births ~ Time + Day + BLY, dist="poisson", data = myDataFrame) #zero inlation model
summary(ziResults)
ziCount <- predict(ziResults, myDataFrame) #zero inflated count
myDataFrame <- cbind(myDataFrame,ziCount) #concatenate calculated count to myDataFrame
The results are as follows
zeroinfl(formula = Births ~ Time + Day + BLY, data = myDataFrame, dist = "poisson")
Pearson residuals:
Min 1Q Median 3Q Max
-1.05159 -0.59996 -0.01862 0.49214 1.87321
Count model coefficients (poisson with log link):
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.51333 0.92356 1.639 0.101
TimeNight -0.01729 0.29613 -0.058 0.953
TimePM 0.04214 0.29228 0.144 0.885
DayWend -0.05728 0.24285 -0.236 0.814
BLY 0.01384 0.18458 0.075 0.940
Zero-inflation model coefficients (binomial with logit link):
Estimate Std. Error z value Pr(>|z|)
(Intercept) -14.472 18736.205 -0.001 0.999
TimeNight -20.209 25675.676 -0.001 0.999
TimePM -20.310 25837.885 -0.001 0.999
DayWend 20.249 18736.200 0.001 0.999
BLY -1.270 3.015 -0.421 0.674
Number of iterations in BFGS optimization: 17
Log-likelihood: -38.26 on 10 Df
> ziCount <- predict(ziResults, myDataFrame) #zero inflated count
> myDataFrame <- cbind(myDataFrame,ziCount) #concatenate calculated count to myDataFrame
> myDataFrame
Time Day BLY Births glmCount ziCount
1 PM Wday 4.5 6 5.140809 5.041806
2 PM Wend 5.6 4 4.500904 4.834177
3 Night Wday 6.0 3 5.070451 4.850580
4 PM Wday 5.9 8 5.368030 5.140475
5 Night Wday 5.2 3 4.946672 4.797157
6 AM Wend 5.8 4 3.635237 3.859393
7 Night Wend 5.9 7 4.277743 4.574203
8 AM Wend 5.0 0 3.546494 2.938806
9 Night Wday 5.5 4 4.992731 4.817121
10 Night Wday 4.1 6 4.781396 4.724659
11 PM Wday 5.4 3 5.285749 5.105017
12 Night Wday 4.2 5 4.796190 4.731204
13 PM Wday 4.7 3 5.172670 5.055785
14 AM Wday 4.4 4 4.113764 4.827064
15 AM Wend 4.7 2 3.513777 2.507346
16 Night Wend 4.8 5 4.134816 4.505074
17 PM Wend 4.8 6 4.391029 4.780934
18 PM Wday 4.5 5 5.140809 5.041806
19 AM Wday 5.0 9 4.190728 4.867325
Finally, in step 2c, the results from the Generalized Linear and Zero Inflated models are compared, using the Vuong Test. The Spearman's Correlation Coefficient is used for comparison
# Step 2c: Comparison of models vuong(glmResults, ziResults)# comparing the two models cor.test( ~ as.numeric(Births) + as.numeric(glmCount), data=myDataFrame, method = "spearman",exact=FALSE) cor.test( ~ as.numeric(Births) + as.numeric(ziCount), data=myDataFrame, method = "spearman",exact=FALSE) cor.test( ~ as.numeric(glmCount) + as.numeric(ziCount), data=myDataFrame, method = "spearman",exact=FALSE)The results are as follows
> vuong(glmResults, ziResults)# comparing the two models
Vuong Non-Nested Hypothesis Test-Statistic:
(test-statistic is asymptotically distributed N(0,1) under the
null that the models are indistinguishible)
-------------------------------------------------------------
Vuong z-statistic H_A p-value
Raw -0.7671979 model2 > model1 0.221482
AIC-corrected 0.9441481 model1 > model2 0.172547
BIC-corrected 1.7522791 model1 > model2 0.039863
>
> cor.test( ~ as.numeric(Births) + as.numeric(glmCount), data=myDataFrame, method = "spearman",exact=FALSE)
Spearman's rank correlation rho
data: as.numeric(Births) and as.numeric(glmCount)
S = 976.91, p-value = 0.559, rho = 0.1430578
> cor.test( ~ as.numeric(Births) + as.numeric(ziCount), data=myDataFrame, method = "spearman",exact=FALSE)
S = 896.89, p-value = 0.3807, rho = 0.2132539
> cor.test( ~ as.numeric(glmCount) + as.numeric(ziCount), data=myDataFrame, method = "spearman",exact=FALSE)
S = 178.16, p-value = 5.647e-06, rho = 0.8437226
The results of the zero inflated analysis presents coefficients for combined Poisson and Binmomial distributions to be used in estimating the counts. The method of calculation using these coefficients is by numerical approximation, and too complex to be explained here. Those interested in numerical methods should consult the references provided.
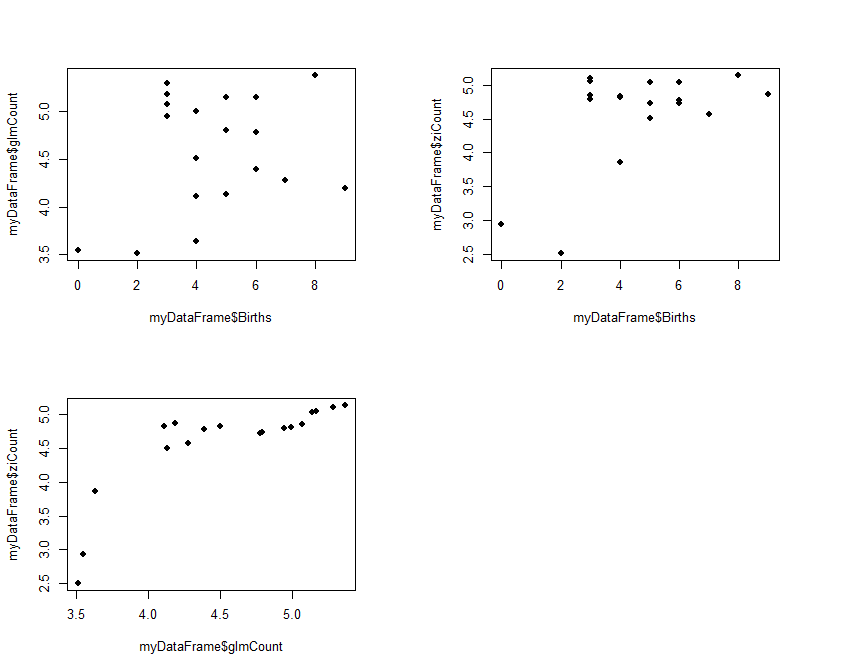
The results (ziCount) are then compared with those from the generalized linear model (glmCount) using the Voung test. The test produced by the basic calculation is labelled Raw, AIC has a correction by Akaike's information criterion, and BIC has a correction by the Bayesian information criterion. In practice they are different methods of calculating the same thing. One way of interpreting the 3 results is to assume that the two models are different if any of the 3 comparisons is statistically significant, as is the case with the current example data. The 3 Spearman's correlations test how closely the two estimates related to the original outcome, and to each other. The results shows that both performed poorly with the reference outcome (Births:glmCount, ρ=0.14; Births:ziCount, ρ=0.21; both not statistically significant). The two estimates however are closely but not perfectly correlated to each other (glmCount:ziCount, ρ=0.84, p<0.0001). Such poor results are not surprising, given that the data is randomly generated by the computer and the sample size very small. It merely serves to demonstrate the numerical methods. Step 3 produces 3 plots in the same view, each a scatterplot for a pair in the 3 counts of Births, glmCont, and ziCount. The plot is optional, but allows the analyst to examine and compare the different results.
#Step 3: Plot fitted values
old.par <- par(mfrow=c(2,2))
par(pin=c(3, 2)) # set plotting window to 4.2x3 inches
plot(x = myDataFrame$Births, # Births on the x axis
y = myDataFrame$glmCount, # glmCount on the y axis
xlim = c(min(myDataFrame$Births),max(myDataFrame$Births)),
ylim = c(min(myDataFrame$glmCount),max(myDataFrame$glmCount)),
pch = 16) # size of dot
plot(x = myDataFrame$Births, # Births on the x axis
y = myDataFrame$ziCount, # ziCount on the y axis
xlim = c(min(myDataFrame$Births),max(myDataFrame$Births)),
ylim = c(min(myDataFrame$ziCount),max(myDataFrame$ziCount)),
pch = 16) # size of dot
plot(x = myDataFrame$glmCount, # Births on the x axis
y = myDataFrame$ziCount, # ziCount on the y axis
xlim = c(min(myDataFrame$glmCount),max(myDataFrame$glmCount)),
ylim = c(min(myDataFrame$ziCount),max(myDataFrame$ziCount)),
pch = 16) # size of dot
par(old.par)
Please note that, in practice, only one of the counts need to be estimated, as by this stage a decision whether glm or zi should be used has been made. The results are presented as follows
# Step 4: Test coefficients on new data set
newDat = ("
Time Day BLY
PM Wday 4.5
Night Wday 6.0
AM Wend 5.8
")
newDataFrame <- read.table(textConnection(newDat),header=TRUE)
summary(newDataFrame)
newGlmCount <- predict(glmResults, newdata=newDataFrame, type='response')
newZiCount <- predict(ziResults, newdata=newDataFrame, type='response')
newDataFrame <- cbind(newDataFrame, newGlmCount, newZiCount)
newDataFrame
The results are as follows
Time Day BLY newGlmCount newZiCount 1 PM Wday 4.5 5.140809 5.041806 2 Night Wday 6.0 5.070451 4.850580 3 AM Wend 5.8 3.635237 3.859393Please note the following
#Step 5: Optional saving and loading of coefficients
#save(glmResults, file = "PoissonGlmResults.rda") #save glm results to rda file
#load("PoissonGlmResults.rda") #load the glm rda file
#save(ziResults, file = "PoissonZiResults.rda") #save zi results to rda file
#load("PoissonZiResults.rda") #load the zi rda file
Step 5 is optional, and in fact commented out, and included as a template only. It allows the results of analysis to be stored as a .rda file, that can be reloaded on a different occasion and used on a different dataset. Please note the following
Contents of C:72
Contents of D:73
Contents of E:74
Contents of H:7
Explanation
R Code
C
D
E
Contents of Explanation:80
Contents of R Code:81
Contents of C:82
Contents of D:83
Contents of E:84
Contents of I:8
Contents of J:9
Contents of K:10
|